A milestone moment for team eDyNAmiC
A milestone moment for team eDyNAmiC and the ecDNA field.
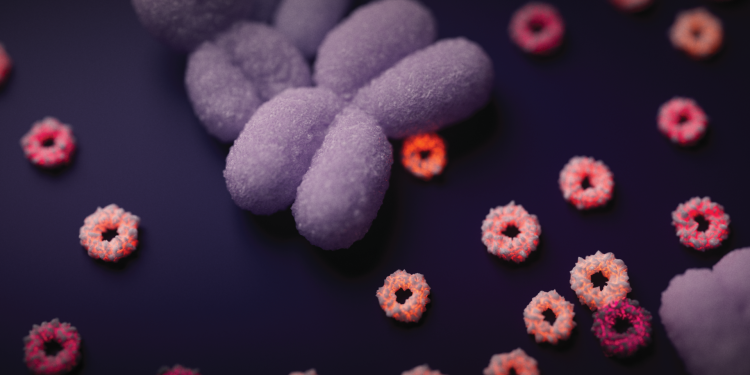
Today team eDyNAmiC and collaborators published three papers in Nature, demonstrating the prevalence and importance of extrachromosomal DNA (ecDNA) in cancer, and going all the way from fundamental mechanisms through to identifying an actionable target that's already being tested in the clinic.
While ecDNA, then known as double minutes, was first observed in tumour cells back in the 1960s, it was thought to be very rare. For decades it received little attention. So how did we get here? And where does team eDyNAmiC come in?
eDyNAmiC team lead Paul Mischel initially trained as a medical doctor, motivated by losing his father to cancer at the age of 14. But after finishing his training he quickly realised it wouldn’t be enough, so at 34 he picked up a pipette for the first time.
‘I became so convinced of the importance of precision medicine after the sequencing of the human genome,’ reflects Paul. But his lab was studying glioblastoma, and trying to understand why patients with highly amplified epidermal growth factor receptor (EGFR) weren’t responding to EGFR inhibitors. ‘Their genome says they should respond, but they don't. I became very frustrated. I was publishing very nice papers. But the patients weren't getting better.’
With the advent of single cell technologies, they began to explore the role of tumour heterogeneity and clonal evolution. In response to EGFR inhibitors, the tumours changed the way the clonal evolution model predicted, selecting for cells that were more resistant, but at a much faster rate. And that wasn’t all.
‘Every single cell in the tumour you could put into a dish or a mouse and the surprise was it gave rise to the original tumour with its full spectrum of heterogeneity. That made no sense by classical genetics,’ explains Paul.
That made them really take a step back, and question all of their assumptions.
‘Here's the map. But what if the map is actually wrong?’ speculates Paul, referring to the map of the human genome.
As genome resolution had increased with improved sequencing technologies, spatial information was actually inferred based on the reference genome - that of a normal cell. This meant there had been an implicit assumption that genes are found in a cancer cell in the same location as a normal cell, but nobody had looked.
So they went back to basics. By using metaphase spreads they demonstrated that the amplified EGFR was not where it should have been on chromosome 7; it was all extrachromosomal. And further, with treatment it all disappeared, re-appearing on ecDNA once the drug was removed, finally alluding to how these tumours were evading treatment. They published their findings in Science in 2014.
‘There was a colossal scratching of heads,’ reflects Paul, ‘and people saying, "well, I guess glioblastoma is an odd genomically unstable tumour. Very nice, but a rarity, of unknown significance."’
It made Paul and others realise that a more thorough understanding of where our oncogenes are amplified in cancer was needed.
It was then that Paul began working with Vineet Bafna, who had previously been involved in sequencing the human genome. Vineet would also later become a key part of Cancer Grand Challenges team eDyNAmiC. The two realised that when they went back to the literature, it was unclear how you actually got so many copies of oncogenes, ‘There was Barbara McClintock's breakage-fusion-bridge work, but that was pretty much it,’ says Paul.
Vineet’s lab developed AmpliconArchitect, which would become critical for the team’s future work, to allow the detection of ecDNA from whole genome sequencing data. Together, by combining the old chromosomal imaging approaches with their newly developed pipelines, looking at 17 different cancer types, they showed that ecDNA was much more widespread than previously thought, found in approximately half the cancer types. Critically they found that many amplified oncogenes were found on ecDNA.
When they published this work in 2017 in Nature, more people started to pay attention, but a lot of people still didn’t know what to do with these findings.
Paul visited Stanford to present this work (his lab was located at UCSD at the time), and it was there he met Howard Chang.
Howard’s lab had become interested in the chromatin structure in cancer cells and had previously developed the game changing ATAC-seq method. When they used ATAC-seq to map the chromatin accessibility landscape of human cancer, they surprisingly found very large tracts of continuously open chromatin. The signal was so large for these areas that they had to mask them out.
But if we looked at the underlying sequence, these were some of the most important cancer-causing genes. It was then that we knew there was something very special going on and that we have to get to the bottom of it.
This turned out to be extrachromosomal DNA.
After Paul’s talk, the two met in Howard’s office, and realised they were coming to the same conclusion. ecDNA was not only allowing basic changes in DNA copy number but also profound epigenetic dysregulation.
Together with Vineet and their collaborators, they published their findings in Nature in 2019, showing the structure and epigenetic content of ecDNA, demonstrating it promotes accessible chromatin, allowing high oncogene expression and informing on function.
By now people were really starting to take notice and realise the scale of the problem.
Also in 2019, Cancer Grand Challenges was setting out to define our third round of challenges, working with the global research community to identify the biggest questions in cancer research. And this time ecDNA was on our radar.
In 2020, Cancer Grand Challenges put the extrachromosomal DNA challenge to the research community.
Paul immediately jumped into action. Aside from thinking about the scientific interests and expertise he wanted to include in the team, it was critical he assembled people with the right motivation to drive the team forward, researchers and advocates alike. 'We are committed to doing this together and I'm actively working with you. But ultimately, the reason we're all doing this isn't for your good or my good. It's for patients' good. And so that was really the spirit and the motivation of how we decided to do it,’ reflects Paul.
Howard and Vineet were already on board.
The growing team anticipated that a large number of ecDNA proteins of interest wouldn’t fall in traditionally druggable classes, so chemist Ben Cravatt was quickly enlisted.
And they had to have Mariam Jamal-Hanjani join them. Much of her pioneering work had centred around trying to understand cancer evolution, where Mariam emphasises, ‘extrachromosomal DNA must play a role.’
Importantly their goals were already aligned. ‘We always come back to the human data and the patient at the heart of the research,’ says Mariam. Mariam runs PEACE, a national autopsy programme in the UK, which crucially enables the collection of tissue from all the different anatomical sites of metastases, to help understand why patients develop drug resistance and ultimately die of metastatic cancer. Together with Charlie Swanton, Mariam also set up TRACERx, which follows lung cancer patients and allows longitudinal genomics throughout the course of their disease.
‘Now, these are not mechanistic studies,’ reminds Mariam. 'They're correlative, but they are human studies, and that's incredibly important to inform both forward and reverse translation - the work that's done preclinically, in vivo and in vitro.’
The star-studded final roster ensured the team could come at the problem from all angles, and critically included patient advocates, keeping the team focused on bringing hope to patients.
As soon as they were established as eDyNAmiC, even before being funded, the researchers were sharing information, analysing data, and working as a team. In 2022 team eDyNAmiC was funded by CRUK and the NCI through Cancer Grand Challenges to tackle the ecDNA challenge.
In the meantime, together with collaborators, they had already published work led by Howard in Nature, showing that ecDNA molecules come together in hubs, with enhancers and genes on different molecules cooperating to drive oncogene expression.
The team and colleagues quickly went on to detail how random inheritance of ecDNA results in extreme intratumoural heterogeneity and allows tumours to dynamically adapt and evolve. They then highlighted the presence of ecDNAs also at early stages, potentially driving transformation in oesophageal cancer.
Now by analysing the genomes from close to 15,000 patients from the Genomics England Cohort, in work co-led by collaborator Charlie Swanton with Mariam and Paul, the team has shown just how prevalent ecDNA is, present in 17.1% of samples. Looking broadly pan-cancer across 39 different tumour types, the presence of ecDNA was associated with tumour stage, treatment, metastases and decreased survival. And the team showed that ecDNA doesn’t just amplify oncogenes, but may play a previously unanticipated role in immune evasion. ‘We see immunomodulatory genes, and genomic regulatory elements, so there's more that ecDNA may contribute,’ says Mariam.
‘Once the cancer cells have ecDNA, they have access to the entire genome,’ poses Howard, so whatever the selective pressure, they can select for genes to give them an advantage. So, under pressure from the immune system they access genes that confer immune escape.
Paul was initially surprised by the sheer diversity of ecDNA elements they detected, but now reflects, ‘The whole mechanism is actually about diversification.’
With regard to the ecDNAs carrying just regulatory elements, such as promoters and enhancers, from the evolutionary perspective Howard thinks of these as altruistic oncogenes: ‘They have no capacity to directly cause any protein production. They can only act indirectly through their partners, other ecDNA molecules. So this is a really new concept in cancer biology.’
The team’s hope is that people will be inspired by this resource, as they have been, drawing even more people to the field who can leverage these findings for their own research.
But how do these altruistic genes ensure they are inherited with their oncogene partners, so they actually have an oncogene to regulate?
By delving deeper the team made the surprising discovery that ecDNAs which cooperate to boost oncogene expression, can actually be coordinately inherited, going against Mendelian rules of inheritance. This is achieved via continued transcription at the start of mitosis, and proximity of the ecDNA molecules, allowing the maintenance of productive ecDNA interactions amidst random segregation.
ecDNA plays by the rules of Charles Darwin but doesn't play by the rules of Gregor Mendel.
Howard likens the mechanism to playing poker: ’If you get some nice cards, imagine how awesome it would be if the next draw you get some of the great cards again, over and over, you're going to keep winning.’
Despite this progress, the question remained of whether ecDNA would be therapeutically actionable.
Going back to Howard’s initial observation that led him into the field, the team wondered whether they could turn this transcriptional advantage of open chromatin into a vulnerability. Upon further exploration they found that ecDNA is not just excessively transcribing protein coding sequences, but it’s transcribing everything. And indeed, this pervasive transcription is creating a problem, leading to increased transcription replication conflict. This stress can be exacerbated by inhibiting CHK1, which leads to preferential death of ecDNA-containing tumour cells.
Howard describes it as a synthetic lethality of excess: ‘Rather than just trying to hit the cancer cells as hard as you can, this strategy is trying to use their own excess, their own momentum against them.’
This approach is already being tested in clinical trials by Boundless Bio*, both as monotherapy and in combination with targeting the oncogenes encoded by the ecDNA.
Paul hopes the team’s findings have the potential to change the course of the lives of patients who are currently being left behind - people perhaps, like his father. By preventing ecDNA from evolving, and taking it completely out of play, he hopes precision medicine could become a success for these patients.
Howard credits the team’s patient advocates with getting them this far, this quickly, already touching on therapeutics: ‘They’ve really influenced our teams thinking along the way.’
And the team is far from done.
By utilising the PEACE and TRACERx** datasets, Paul emphasises, ‘We have a remarkably important opportunity to learn from patients over space and time, to learn how to track ecDNA and to diagnose it earlier.’
The team is working to understand how ecDNA shuts off the immune system, and how we can bring it back into play.
eDyNAmiC also continues to disentangle the molecular mechanisms of how ecDNA goes through the fundamental processes of transcription, replication and repair - with the hope that each of these processes may be different and targetable. As Howard puts it, ‘That's all happening on the chromatin template and for ecDNA that chromatin is different. So every one of these steps could have differences that we can perhaps exploit for therapy.’ As the team discovers new potential targets, it is leveraging Ben Cravatt’s unique chemical approaches to drug the undruggable.
For Paul and the team, Cancer Grand Challenges is the opportunity to make a difference for people with the most difficult types of cancer: ‘Allowing us to do it in a way that would just be impossible under any other system. And we've just begun. We see a different future in sight if we can get this right.’
Learn more about this milestone moment for eDyNAmiC
-------------------------
Papers:
Find out more about team eDyNAmiC. Through Cancer Grand Challenges team eDyNAmiC is funded by Cancer Research UK and the National Cancer Institute, with generous support to Cancer Research UK from Emerson Collective and The Kamini and Vindi Banga Family Trust.
*Boundless Bio is a Nasdaq-listed company, founded in 2018 by some members of the team.
**PEACE and TRACERx are supported by Cancer Research UK.
Article written by Rebecca Eccles with thanks to Paul Mischel, Howard Chang and Mariam Jamal-Hanjani .
Affiliations:
Professor Paul Mischel is a Professor of Pathology and, by courtesy, of Neurosurgery, and the Fortinet Founders Professor at the Stanford School of Medicine. He is also Vice Chair for Research in the Department of Pathology, and a Sarafan ChEM-H Institute Scholar.
Professor Vineet Bafna is Professor of Computer Science and Engineering at the Halicioglu Data Science Institute at University of California, San Diego.
Professor Howard Chang is a Professor of Dermatology and Genetics and the Virginia and D.K. Ludwig Professor of Cancer Research at the Stanford School of Medicine and a Howard Hughes Medical Institute Investigator.
Professor Ben Cravatt is Professor and Gilula Chair of Chemical Biology in the Department of Chemistry at The Scripps Research Institute.
Professor Mariam Jamal-Hanjani is Professor of Cancer Genomics and Metastasis at the Cancer Research UK Lung Cancer Centre of Excellence at University College London (UCL) Cancer Institute. She is also an Honorary Medical Oncology Consultant in Translational Lung Oncology with the UCL Hospitals NHS Trust.
Professor Charles Swanton is the Deputy Clinical Director at the Francis Crick Institute and Chair in Personalised Medicine at UCL. He is also the Royal Society Napier Professor in Cancer and thoracic medical oncologist at UCL Hospitals, co-director of the Cancer Research UK (CRUK) Lung Cancer Centre of Excellence, and Chief Clinician of Cancer Research UK.
Hero image: Artistic visualisation of extrachromosomal DNA. The large, purple rod-shaped object is a chromosome. The smaller orange-red circles are the extrachromosomal DNA. Credit: Jeroen Claus.
A milestone moment for team eDyNAmiC and the ecDNA field.
Q&A with eDyNAmiC future leader Chris Bailey.